Laboratory information:Contact information:
Sampsa Hautaniemi Biomedicum Helsinki, B524b P.O.Box 63 (Haartmaninkatu 8) 00014 UNIVERSITY OF HELSINKI phone +358 9 191 25419 fax +358 9 191 25610 firstname.lastname@helsinki.fi |
Research projects
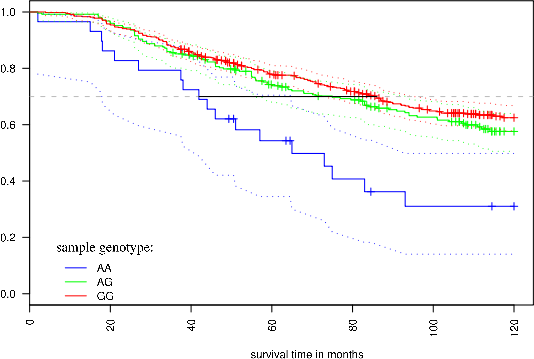
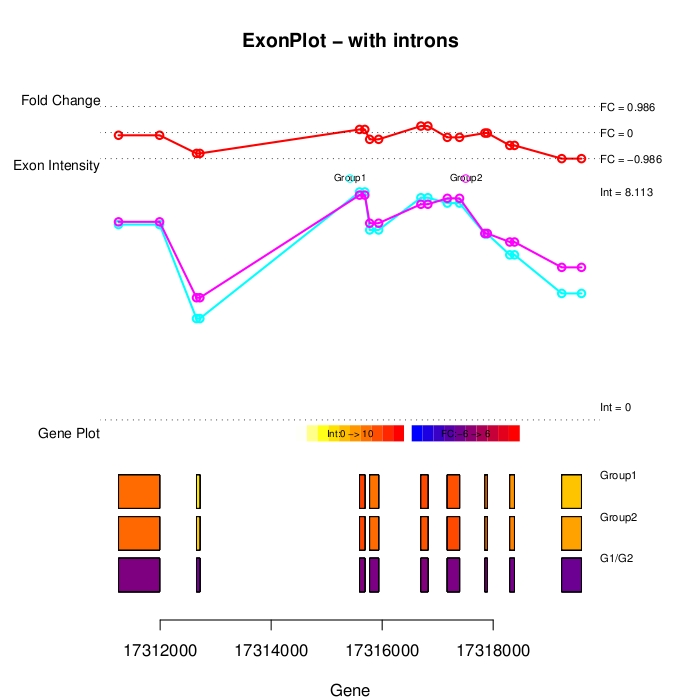
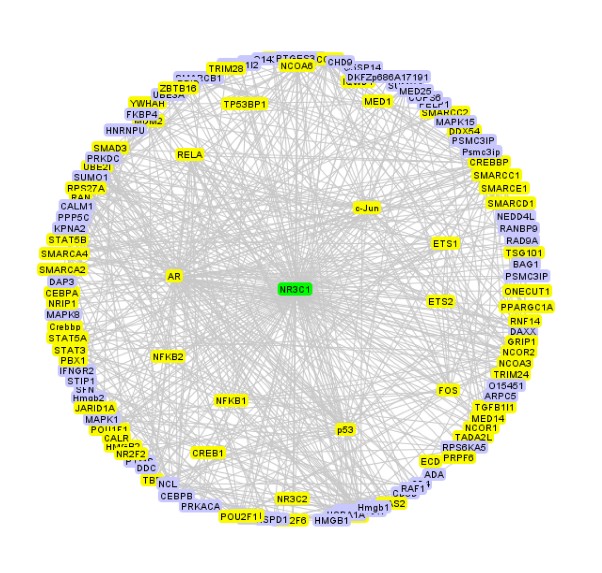
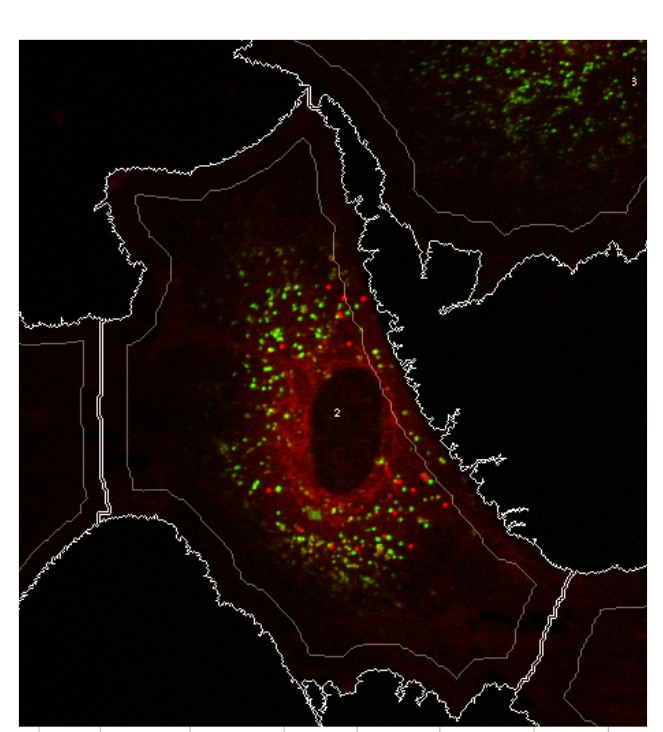
Cancer genetic data mining Genetic variance is a major factor in several diseases, such as cancer. One of the most frequent type of genetic variation is single nucleotide polymorphism (SNP), i.e., a change of one nucleotide to another. There are more than 10 million in SNPs in human genome, and these SNPs are typically highly conserved within population making SNPs excellent genotypic markers for medical research. With current microarray technology (SNP-arrays) it is possible to measure up to one million SNPs in any human tissue sample. Our objectives are to develop computational tools to efficiently analyze SNP-array data in cancers and to identify combinatorially acting genes. The underlying principle of our efforts is to maximize sensitivity (i.e. maximize the probability that disease associated variants, if any, are found) and then integrate data from biological databases and published studies to exclude false positives. High-throughput data analysis In many diseases, genes or proteins playing key roles in disease progression, drug resistance and decreased survival are poorly known. High-throughput methods have become standard technology in identifying genes and proteins that are most likely involved in disease initiation, progression and drug resistance. We have a strong track-record in experimental design, preprocessing and analyzing data from a number of high-throughput technologies such as gene, exon, SNP, copy-number microarrays, mass spectrometry and Illumina Solexa deep-sequencing platform. Our objective is to translate the raw high-throughput data into experimentally testable predictions that increase knowledge on disease mechanisms and effective means of treatment. Data integration Data integration is the key element in translating high-throughput data into trustworthy predictions. Integration is hindered by two challenges: (i) data come from different sources in various (incompatible) formats and (ii) software for bioinformatics often does not work as promised and/or is challenging to use. To overcome these challenges we have developed an open source component-based workflow framework called Anduril for data analysis and integration. The components can be implemented with any language, such as Java, Python, MATLAB, C++, Weka and R. Anduril automatically produces reports consisting of all methods, their parameters, figures, tables and so on. We currently provide components for expression, SNP, ChIP-chip and exon microarray analysis, and third parties can develop new components. Implemented database connections include Ensembl, KEGG, PINA and others.Signaling pathway modeling The vast majority of diseases are appreciated to be "complex"; They arise from alterations within multiple molecular regulatory pathways. Signaling pathways represent a crucial domain for pathological dysregulation as they contain forward and reverse feedback cascades that can act as signal amplifiers, transmitters, or distributors to a multitude of highly-connected protein nodes across numerous pathways within a network. We develop and use mechanistic (e.g. differential equation) and data-driven (e.g. decision tree) based modeling approaches to characterize the impact of genetic variation, gene/protein expression patterns, and inhibitors on signaling pathways. At the moment, our focus is on modeling EGFR, NF-kB and Fas pathways in cancer cells. Image analysis Due to advances in cell imaging, image processing is getting increasingly important in systems biology. Our efforts focus on efficient and automated analysis of fluorescence imaging, live cell imaging and using the information extracted from images in statistical analysis. We belong to the FLUODIAMON consortium funded by EU FP7. The overall objective of FLUODIAMON is to develop and validate a quantitative, minimally invasive diagnostic tool for early and conclusive detection, diagnosis and monitoring of disease and disease progression of breast and prostate cancer, with negligible sampling-related side-effects. We are responsible for bioinformatics analysis in the consortium. |